Cell nucleus (W)
Cell nucleus (W)
|
| |
|
In cell biology, the nucleus (pl. nuclei; from Latin nucleus or nuculeus, meaning kernel or seed) is a membrane-bound organelle found in eukaryotic cells. Eukaryotes usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix (which includes the nuclear lamina), a network within the nucleus that adds mechanical support, much like the cytoskeleton supports the cell as a whole.
The cell nucleus contains all of the cell’s genome, except for a small fraction of mitochondrial DNA, organized as multiple long linear DNA molecules in a complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene expression—the nucleus is, therefore, the control center of the cell.
Because the nuclear envelope is impermeable to large molecules, nuclear pores are required to regulate nuclear transport of molecules across the envelope. The pores cross both nuclear membranes, providing a channel through which larger molecules must be actively transported by carrier proteins while allowing free movement of small molecules and ions. Movement of large molecules such as proteins and RNA through the pores is required for both gene expression and the maintenance of chromosomes.
Although the interior of the nucleus does not contain any membrane-bound subcompartments, its contents are not uniform, and a number of nuclear bodies exist, made up of unique proteins, RNA molecules, and particular parts of the chromosomes. The best-known of these is the nucleolus, which is mainly involved in the assembly of ribosomes. After being produced in the nucleolus, ribosomes are exported to the cytoplasm where they translate mRNA. |
|
| |
| |
History
|
History
History (W)
The nucleus was the first organelle to be discovered. What is most likely the oldest preserved drawing dates back to the early microscopist Antonie van Leeuwenhoek (1632–1723). He observed a "lumen", the nucleus, in the red blood cells of salmon. Unlike mammalian red blood cells, those of other vertebrates still contain nuclei.
The nucleus was also described by Franz Bauer in 1804 and in more detail in 1831 by Scottish botanist Robert Brown in a talk at the Linnean Society of London. Brown was studying orchids under the microscope when he observed an opaque area, which he called the "areola" or "nucleus", in the cells of the flower's outer layer.
He did not suggest a potential function. In 1838, Matthias Schleiden proposed that the nucleus plays a role in generating cells, thus he introduced the name "cytoblast" ("cell builder"). He believed that he had observed new cells assembling around "cytoblasts". Franz Meyen was a strong opponent of this view, having already described cells multiplying by division and believing that many cells would have no nuclei. The idea that cells can be generated de novo, by the "cytoblast" or otherwise, contradicted work by Robert Remak (1852) and Rudolf Virchow (1855) who decisively propagated the new paradigm that cells are generated solely by cells ("Omnis cellula e cellula"). The function of the nucleus remained unclear.
Between 1877 and 1878, Oscar Hertwig published several studies on the fertilization of sea urchin eggs, showing that the nucleus of the sperm enters the oocyte and fuses with its nucleus. This was the first time it was suggested that an individual develops from a (single) nucleated cell. This was in contradiction to Ernst Haeckel's theory that the complete phylogeny of a species would be repeated during embryonic development, including generation of the first nucleated cell from a "monerula", a structureless mass of primordial protoplasm ("Urschleim"). Therefore, the necessity of the sperm nucleus for fertilization was discussed for quite some time. However, Hertwig confirmed his observation in other animal groups, including amphibians and molluscs. Eduard Strasburger produced the same results for plants in 1884. This paved the way to assign the nucleus an important role in heredity. In 1873, August Weismann postulated the equivalence of the maternal and paternal germ cells for heredity. The function of the nucleus as carrier of genetic information became clear only later, after mitosis was discovered and the Mendelian rules were rediscovered at the beginning of the 20th century; the chromosome theory of heredity was therefore developed. |

|
|
|
|
| |
Structures
|
Structures
Structures (W)
The nucleus is the largest organelle in animal cells. In mammalian cells, the average diameter of the nucleus is approximately 6 micrometres (µm), which occupies about 10% of the total cell volume. The contents of the nucleus are held in the nucleoplasm similar to the cytoplasm in the rest of the cell. The fluid component of this is termed the nucleosol, similar to the cytosol in the cytoplasm.
In most types of granulocyte, a white blood cell, the nucleus is lobated and can be bi-lobed, tri-lobed or multi-lobed.
The dynamic behaviour of structures in the nucleus, such as the nuclear rotation that occurs prior to mitosis, can be visualized using label-free live cell imaging. |
| |
|
|
|
|
Nuclear envelope and pores
Nuclear envelope and pores (W)
The nuclear envelope, otherwise known as nuclear membrane, consists of two cellular membranes, an inner and an outer membrane, arranged parallel to one another and separated by 10 to 50 nanometres (nm). The nuclear envelope completely encloses the nucleus and separates the cell's genetic material from the surrounding cytoplasm, serving as a barrier to prevent macromolecules from diffusing freely between the nucleoplasm and the cytoplasm. The outer nuclear membrane is continuous with the membrane of the rough endoplasmic reticulum (RER), and is similarly studded with ribosomes. The space between the membranes is called the perinuclear space and is continuous with the RER lumen.
Nuclear pores, which provide aqueous channels through the envelope, are composed of multiple proteins, collectively referred to as nucleoporins. The pores are about 125 million daltons in molecular weight and consist of around 50 (in yeast) to several hundred proteins (in vertebrates). The pores are 100 nm in total diameter; however, the gap through which molecules freely diffuse is only about 9 nm wide, due to the presence of regulatory systems within the center of the pore. This size selectively allows the passage of small water-soluble molecules while preventing larger molecules, such as nucleic acids and larger proteins, from inappropriately entering or exiting the nucleus. These large molecules must be actively transported into the nucleus instead. The nucleus of a typical mammalian cell will have about 3000 to 4000 pores throughout its envelope, each of which contains an eightfold-symmetric ring-shaped structure at a position where the inner and outer membranes fuse. Attached to the ring is a structure called the nuclear basket that extends into the nucleoplasm, and a series of filamentous extensions that reach into the cytoplasm. Both structures serve to mediate binding to nuclear transport proteins.
Most proteins, ribosomal subunits, and some DNAs are transported through the pore complexes in a process mediated by a family of transport factors known as karyopherins. Those karyopherins that mediate movement into the nucleus are also called importins, whereas those that mediate movement out of the nucleus are called exportins. Most karyopherins interact directly with their cargo, although some use adaptor proteins. Steroid hormones such as cortisol and aldosterone, as well as other small lipid-soluble molecules involved in intercellular signaling, can diffuse through the cell membrane and into the cytoplasm, where they bind nuclear receptor proteins that are trafficked into the nucleus. There they serve as transcription factors when bound to their ligand; in the absence of a ligand, many such receptors function as histone deacetylases that repress gene expression. |
| |
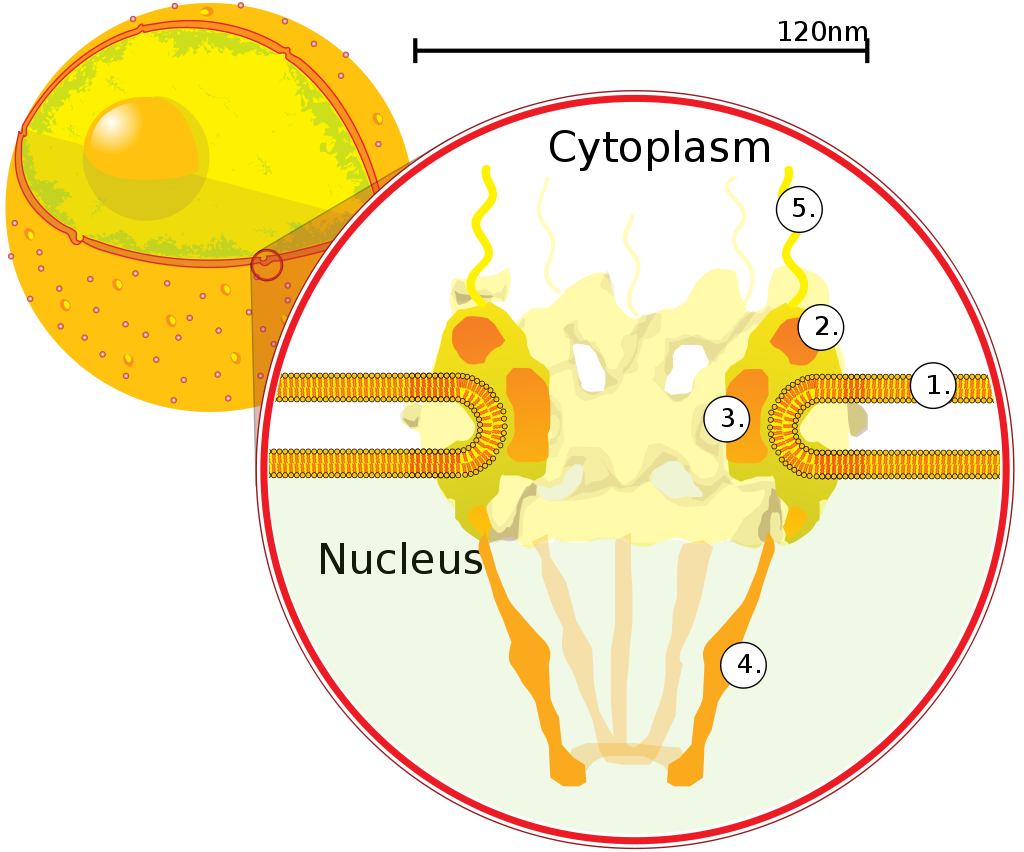
A cross section of a nuclear pore on the surface of the nuclear envelope (1). Other diagram labels show (2) the outer ring, (3) spokes, (4) basket, and (5) filaments. |
|
|

|
|
|
|
Nuclear lamina
Nuclear lamina (W)
In animal cells, two networks of intermediate filaments provide the nucleus with mechanical support: The nuclear lamina forms an organized meshwork on the internal face of the envelope, while less organized support is provided on the cytosolic face of the envelope. Both systems provide structural support for the nuclear envelope and anchoring sites for chromosomes and nuclear pores.
The nuclear lamina is composed mostly of lamin proteins. Like all proteins, lamins are synthesized in the cytoplasm and later transported to the nucleus interior, where they are assembled before being incorporated into the existing network of nuclear lamina.Lamins found on the cytosolic face of the membrane, such as emerin and nesprin, bind to the cytoskeleton to provide structural support. Lamins are also found inside the nucleoplasm where they form another regular structure, known as the nucleoplasmic veil,that is visible using fluorescence microscopy. The actual function of the veil is not clear, although it is excluded from the nucleolus and is present during interphase.Lamin structures that make up the veil, such as LEM3, bind chromatin and disrupting their structure inhibits transcription of protein-coding genes.
Like the components of other intermediate filaments, the lamin monomer contains an alpha-helical domain used by two monomers to coil around each other, forming a dimer structure called a coiled coil. Two of these dimer structures then join side by side, in an antiparallel arrangement, to form a tetramer called a protofilament. Eight of these protofilaments form a lateral arrangement that is twisted to form a ropelike filament. These filaments can be assembled or disassembled in a dynamic manner, meaning that changes in the length of the filament depend on the competing rates of filament addition and removal.
Mutations in lamin genes leading to defects in filament assembly cause a group of rare genetic disorders known as laminopathies. The most notable laminopathy is the family of diseases known as progeria, which causes the appearance of premature aging in its sufferers. The exact mechanism by which the associated biochemical changes give rise to the aged phenotype is not well understood. |

|
|
|
|
Chromosomes
Chromosomes (W)
The cell nucleus contains the majority of the cell's genetic material in the form of multiple linear DNA molecules organized into structures called chromosomes. Each human cell contains roughly two meters of DNA.:405 During most of the cell cycle these are organized in a DNA-protein complex known as chromatin, and during cell division the chromatin can be seen to form the well-defined chromosomes familiar from a karyotype. A small fraction of the cell's genes are located instead in the mitochondria.
There are two types of chromatin. Euchromatin is the less compact DNA form, and contains genes that are frequently expressed by the cell. The other type, heterochromatin, is the more compact form, and contains DNA that is infrequently transcribed. This structure is further categorized into facultative heterochromatin, consisting of genes that are organized as heterochromatin only in certain cell types or at certain stages of development, and constitutive heterochromatin that consists of chromosome structural components such as telomeres and centromeres. During interphase the chromatin organizes itself into discrete individual patches, called chromosome territories.Active genes, which are generally found in the euchromatic region of the chromosome, tend to be located towards the chromosome's territory boundary.
Antibodies to certain types of chromatin organization, in particular, nucleosomes, have been associated with a number of autoimmune diseases, such as systemic lupus erythematosus. These are known as anti-nuclear antibodies (ANA) and have also been observed in concert with multiple sclerosis as part of general immune system dysfunction. |

|
|
|
|
Nucleolus
Nucleolus (W)
The nucleolus is the largest of the discrete densely stained, membraneless structures known as nuclear bodies found in the nucleus. It forms around tandem repeats of rDNA, DNA coding for ribosomal RNA (rRNA). These regions are called nucleolar organizer regions (NOR). The main roles of the nucleolus are to synthesize rRNA and assemble ribosomes. The structural cohesion of the nucleolus depends on its activity, as ribosomal assembly in the nucleolus results in the transient association of nucleolar components, facilitating further ribosomal assembly, and hence further association. This model is supported by observations that inactivation of rDNA results in intermingling of nucleolar structures.
In the first step of ribosome assembly, a protein called RNA polymerase I transcribes rDNA, which forms a large pre-rRNA precursor. This is cleaved into the subunits 5.8S, 18S, and 28S rRNA. The transcription, post-transcriptional processing, and assembly of rRNA occurs in the nucleolus, aided by small nucleolar RNA (snoRNA) molecules, some of which are derived from spliced introns from messenger RNAs encoding genes related to ribosomal function. The assembled ribosomal subunits are the largest structures passed through the nuclear pores.
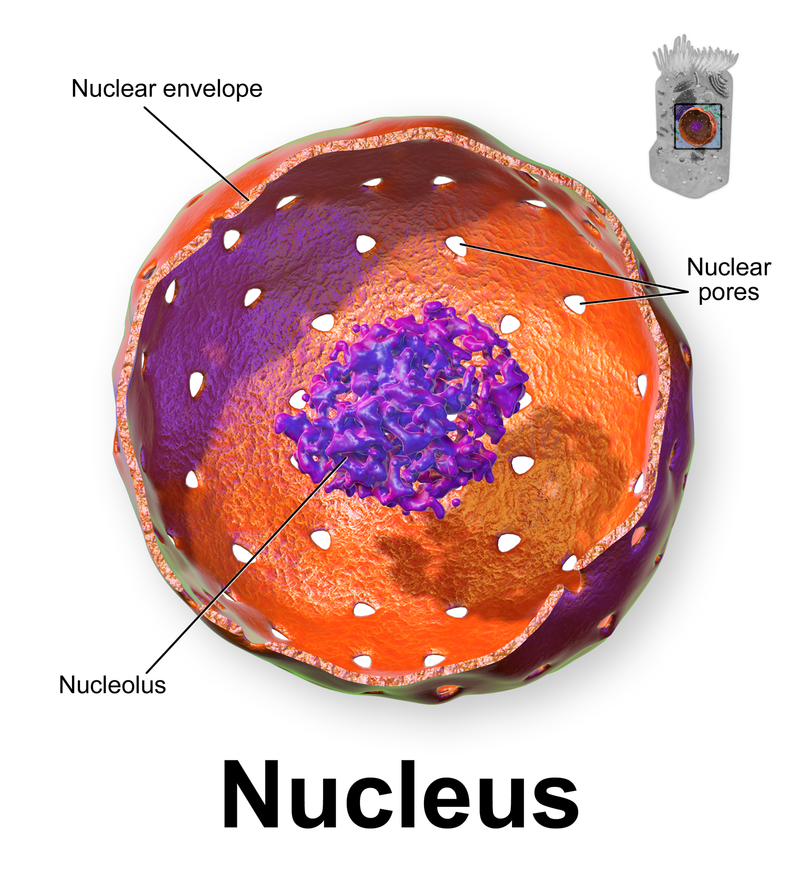
3D rendering of nucleus with location of nucleolus. |
|
|
When observed under the electron microscope, the nucleolus can be seen to consist of three distinguishable regions: the innermost fibrillar centers (FCs), surrounded by the dense fibrillar component (DFC) (that contains fibrillarin and nucleolin), which in turn is bordered by the granular component (GC) (that contains the protein nucleophosmin). Transcription of the rDNA occurs either in the FC or at the FC-DFC boundary, and, therefore, when rDNA transcription in the cell is increased, more FCs are detected. Most of the cleavage and modification of rRNAs occurs in the DFC, while the latter steps involving protein assembly onto the ribosomal subunits occur in the GC. |

|
|
|
|
Other nuclear bodies
Other nuclear bodies (W)
Besides the nucleolus, the nucleus contains a number of other nuclear bodies. These include Cajal bodies, gemini of Cajal bodies, polymorphic interphase karyosomal association (PIKA), promyelocytic leukaemia (PML) bodies, paraspeckles, and splicing speckles. Although little is known about a number of these domains, they are significant in that they show that the nucleoplasm is not a uniform mixture, but rather contains organized functional subdomains.
Other subnuclear structures appear as part of abnormal disease processes. For example, the presence of small intranuclear rods has been reported in some cases of nemaline myopathy. This condition typically results from mutations in actin, and the rods themselves consist of mutant actin as well as other cytoskeletal proteins. |
| |
Subnuclear structure sizes
| Structure name |
Structure diameter |
Ref. |
| Cajal bodies |
0.2–2.0 µm |
[34] |
| Clastosomes |
0.2-0.5 µm |
[35] |
| PIKA |
5 µm |
[36] |
| PML bodies |
0.2–1.0 µm |
[37] |
| Paraspeckles |
0.5–1.0 µm |
[38] |
| Speckles |
20–25 nm |
[36] |
|

|
|
|
|
Cajal bodies and gems
Cajal bodies and gems (W)
A nucleus typically contains between 1 and 10 compact structures called Cajal bodies or coiled bodies (CB), whose diameter measures between 0.2 µm and 2.0 µm depending on the cell type and species. When seen under an electron microscope, they resemble balls of tangled thread and are dense foci of distribution for the protein coilin. CBs are involved in a number of different roles relating to RNA processing, specifically small nucleolar RNA (snoRNA) and small nuclear RNA (snRNA) maturation, and histone mRNA modification.
Similar to Cajal bodies are Gemini of Cajal bodies, or gems, whose name is derived from the Gemini constellation in reference to their close "twin" relationship with CBs. Gems are similar in size and shape to CBs, and in fact are virtually indistinguishable under the microscope. Unlike CBs, gems do not contain small nuclear ribonucleoproteins (snRNPs), but do contain a protein called survival of motor neuron (SMN) whose function relates to snRNP biogenesis. Gems are believed to assist CBs in snRNP biogenesis, though it has also been suggested from microscopy evidence that CBs and gems are different manifestations of the same structure. Later ultrastructural studies have shown gems to be twins of Cajal bodies with the difference being in the coilin component; Cajal bodies are SMN positive and coilin positive, and gems are SMN positive and coilin negative. |

|
|
|
|
PIKA and PTF domains
PIKA and PTF domains (W)
PIKA domains, or polymorphic interphase karyosomal associations, were first described in microscopy studies in 1991. Their function remains unclear, though they were not thought to be associated with active DNA replication, transcription, or RNA processing. They have been found to often associate with discrete domains defined by dense localization of the transcription factor PTF, which promotes transcription of small nuclear RNA (snRNA). |
|
|
|
PML bodies
PML bodies (W)
Promyelocytic leukemia bodies (PML bodies) are spherical bodies found scattered throughout the nucleoplasm, measuring around 0.1–1.0 µm. They are known by a number of other names, including nuclear domain 10 (ND10), Kremer bodies, and PML oncogenic domains. PML bodies are named after one of their major components, the promyelocytic leukemia protein (PML). They are often seen in the nucleus in association with Cajal bodies and cleavage bodies. Pml-/- mice, which are unable to create PML bodies, develop normally without obvious ill effects, showing that PML bodies are not required for most essential biological processes. |
|
|
|
Splicing speckles
Splicing speckles (W)
Speckles are subnuclear structures that are enriched in pre-messenger RNA splicing factors and are located in the interchromatin regions of the nucleoplasm of mammalian cells. At the fluorescence-microscope level they appear as irregular, punctate structures, which vary in size and shape, and when examined by electron microscopy they are seen as clusters of interchromatin granules. Speckles are dynamic structures, and both their protein and RNA-protein components can cycle continuously between speckles and other nuclear locations, including active transcription sites. Studies on the composition, structure and behaviour of speckles have provided a model for understanding the functional compartmentalization of the nucleus and the organization of the gene-expression machinery splicing snRNPs and other splicing proteins necessary for pre-mRNA processing. Because of a cell's changing requirements, the composition and location of these bodies changes according to mRNA transcription and regulation via phosphorylation of specific proteins. The splicing speckles are also known as nuclear speckles (nuclear specks), splicing factor compartments (SF compartments), interchromatin granule clusters (IGCs), and B snurposomes. B snurposomes are found in the amphibian oocyte nuclei and in Drosophila melanogaster embryos. B snurposomes appear alone or attached to the Cajal bodies in the electron micrographs of the amphibian nuclei. IGCs function as storage sites for the splicing factors. |

|
|
|
|
Paraspeckles
Paraspeckles (W)
Discovered by Fox et al. in 2002, paraspeckles are irregularly shaped compartments in the interchromatin space of the nucleus. First documented in HeLa cells, where there are generally 10–30 per nucleus, paraspeckles are now known to also exist in all human primary cells, transformed cell lines, and tissue sections. Their name is derived from their distribution in the nucleus; the "para" is short for parallel and the "speckles" refers to the splicing speckles to which they are always in close proximity.
Paraspeckles sequester nuclear proteins and RNA and thus appear to function as a molecular sponge that is involved in the regulation of gene expression. Furthermore, paraspeckles are dynamic structures that are altered in response to changes in cellular metabolic activity. They are transcription dependent and in the absence of RNA Pol II transcription, the paraspeckle disappears and all of its associated protein components (PSP1, p54nrb, PSP2, CFI(m)68, and PSF) form a crescent shaped perinucleolar cap in the nucleolus. This phenomenon is demonstrated during the cell cycle. In the cell cycle, paraspeckles are present during interphase and during all of mitosis except for telophase. During telophase, when the two daughter nuclei are formed, there is no RNA Pol II transcription so the protein components instead form a perinucleolar cap. |

|
|
|
|
Perichromatin fibrils
Perichromatin fibrils (W)
Perichromatin fibrils are visible only under electron microscope. They are located next to the transcriptionally active chromatin and are hypothesized to be the sites of active pre-mRNA processing. |
|
|
|
Clastosomes
Clastosomes (W)
Clastosomes are small nuclear bodies (0.2–0.5 µm) described as having a thick ring-shape due to the peripheral capsule around these bodies. This name is derived from the Greek klastos, broken and soma, body. Clastosomes are not typically present in normal cells, making them hard to detect. They form under high proteolytic conditions within the nucleus and degrade once there is a decrease in activity or if cells are treated with proteasome inhibitors. The scarcity of clastosomes in cells indicates that they are not required for proteasome function. Osmotic stress has also been shown to cause the formation of clastosomes. These nuclear bodies contain catalytic and regulatory subunits of the proteasome and its substrates, indicating that clastosomes are sites for degrading proteins. |
|
|
|
| |
Function
|
Function
Function (W)
The nucleus provides a site for genetic transcription that is segregated from the location of translation in the cytoplasm, allowing levels of gene regulation that are not available to prokaryotes. The main function of the cell nucleus is to control gene expression and mediate the replication of DNA during the cell cycle.
The nucleus is an organelle found in eukaryotic cells. Inside its fully enclosed nuclear membrane, it contains the majority of the cell's genetic material. This material is organized as DNA molecules, along with a variety of proteins, to form chromosomes. |
|
|
|
Cell compartmentalization
Cell compartmentalization (W)
The nuclear envelope allows the nucleus to control its contents, and separate them from the rest of the cytoplasm where necessary. This is important for controlling processes on either side of the nuclear membrane. In most cases where a cytoplasmic process needs to be restricted, a key participant is removed to the nucleus, where it interacts with transcription factors to downregulate the production of certain enzymes in the pathway. This regulatory mechanism occurs in the case of glycolysis, a cellular pathway for breaking down glucose to produce energy. Hexokinase is an enzyme responsible for the first the step of glycolysis, forming glucose-6-phosphate from glucose. At high concentrations of fructose-6-phosphate, a molecule made later from glucose-6-phosphate, a regulator protein removes hexokinase to the nucleus, where it forms a transcriptional repressor complex with nuclear proteins to reduce the expression of genes involved in glycolysis.
In order to control which genes are being transcribed, the cell separates some transcription factor proteins responsible for regulating gene expression from physical access to the DNA until they are activated by other signaling pathways. This prevents even low levels of inappropriate gene expression. For example, in the case of NF-κB-controlled genes, which are involved in most inflammatory responses, transcription is induced in response to a signal pathway such as that initiated by the signaling molecule TNF-α, binds to a cell membrane receptor, resulting in the recruitment of signalling proteins, and eventually activating the transcription factor NF-κB. A nuclear localisation signal on the NF-κB protein allows it to be transported through the nuclear pore and into the nucleus, where it stimulates the transcription of the target genes.
The compartmentalization allows the cell to prevent translation of unspliced mRNA. Eukaryotic mRNA contains introns that must be removed before being translated to produce functional proteins. The splicing is done inside the nucleus before the mRNA can be accessed by ribosomes for translation. Without the nucleus, ribosomes would translate newly transcribed (unprocessed) mRNA, resulting in malformed and nonfunctional proteins. |

|
|
|
|
Replication
Replication (W)
The main function of the cell nucleus is to control gene expression and mediate the replication of DNA during the cell cycle. It has been found that replication happens in a localised way in the cell nucleus. In the S phase of interphase of the cell cycle; replication takes place. Contrary to the traditional view of moving replication forks along stagnant DNA, a concept of replication factories emerged, which means replication forks are concentrated towards some immobilised 'factory' regions through which the template DNA strands pass like conveyor belts. |
|
|
|
Gene expression
Gene expression (W)
|
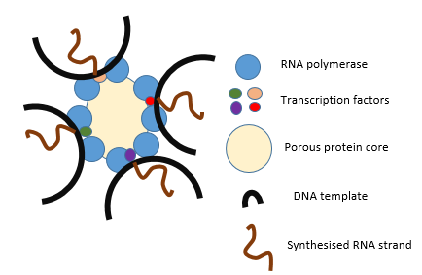
A generic transcription factory during transcription, highlighting the possibility of transcribing more than one gene at a time. The diagram includes 8 RNA polymerases however the number can vary depending on cell type. The image also includes transcription factors and a porous, protein core. |
|
|
| |
|
Gene expression first involves transcription, in which DNA is used as a template to produce RNA. In the case of genes encoding proteins, that RNA produced from this process is messenger RNA (mRNA), which then needs to be translated by ribosomes to form a protein. As ribosomes are located outside the nucleus, mRNA produced needs to be exported.
Since the nucleus is the site of transcription, it also contains a variety of proteins that either directly mediate transcription or are involved in regulating the process. These proteins include helicases, which unwind the double-stranded DNA molecule to facilitate access to it, RNA polymerases, which bind to the DNA promoter to synthesize the growing RNA molecule, topoisomerases, which change the amount of supercoiling in DNA, helping it wind and unwind, as well as a large variety of transcription factors that regulate expression. |
| |
|

|
|
|
|
Processing of pre-mRNA
Processing of pre-mRNA (W)
Newly synthesized mRNA molecules are known as primary transcripts or pre-mRNA. They must undergo post-transcriptional modification in the nucleus before being exported to the cytoplasm; mRNA that appears in the cytoplasm without these modifications is degraded rather than used for protein translation. The three main modifications are 5' capping, 3' polyadenylation, and RNA splicing. While in the nucleus, pre-mRNA is associated with a variety of proteins in complexes known as heterogeneous ribonucleoprotein particles (hnRNPs). Addition of the 5' cap occurs co-transcriptionally and is the first step in post-transcriptional modification. The 3' poly-adenine tail is only added after transcription is complete.
RNA splicing, carried out by a complex called the spliceosome, is the process by which introns, or regions of DNA that do not code for protein, are removed from the pre-mRNA and the remaining exons connected to re-form a single continuous molecule. This process normally occurs after 5' capping and 3' polyadenylation but can begin before synthesis is complete in transcripts with many exons.Many pre-mRNAs can be spliced in multiple ways to produce different mature mRNAs that encode different protein sequences. This process is known as alternative splicing, and allows production of a large variety of proteins from a limited amount of DNA. |

|
|
|
|
| |
Dynamics and regulation
|
Dynamics and regulation
Dynamics and regulation (W)
No text. |
|
|
|
Nuclear transport
Nuclear transport (W)
The entry and exit of large molecules from the nucleus is tightly controlled by the nuclear pore complexes. Although small molecules can enter the nucleus without regulation, macromolecules such as RNA and proteins require association karyopherins called importins to enter the nucleus and exportins to exit. "Cargo" proteins that must be translocated from the cytoplasm to the nucleus contain short amino acid sequences known as nuclear localization signals, which are bound by importins, while those transported from the nucleus to the cytoplasm carry nuclear export signals bound by exportins. The ability of importins and exportins to transport their cargo is regulated by GTPases, enzymes that hydrolyze the molecule guanosine triphosphate (GTP) to release energy. The key GTPase in nuclear transport is Ran, which can bind either GTP or GDP (guanosine diphosphate), depending on whether it is located in the nucleus or the cytoplasm. Whereas importins depend on RanGTP to dissociate from their cargo, exportins require RanGTP in order to bind to their cargo.
Nuclear import depends on the importin binding its cargo in the cytoplasm and carrying it through the nuclear pore into the nucleus. Inside the nucleus, RanGTP acts to separate the cargo from the importin, allowing the importin to exit the nucleus and be reused. Nuclear export is similar, as the exportin binds the cargo inside the nucleus in a process facilitated by RanGTP, exits through the nuclear pore, and separates from its cargo in the cytoplasm.
Specialized export proteins exist for translocation of mature mRNA and tRNA to the cytoplasm after post-transcriptional modification is complete. This quality-control mechanism is important due to these molecules' central role in protein translation. Mis-expression of a protein due to incomplete excision of exons or mis-incorporation of amino acids could have negative consequences for the cell; thus, incompletely modified RNA that reaches the cytoplasm is degraded rather than used in translation. |
| |
|

|
|
|
|
Assembly and disassembly
Assembly and disassembly (W)
During its lifetime, a nucleus may be broken down or destroyed, either in the process of cell division or as a consequence of apoptosis (the process of programmed cell death). During these events, the structural components of the nucleus — the envelope and lamina — can be systematically degraded. In most cells, the disassembly of the nuclear envelope marks the end of the prophase of mitosis. However, this disassembly of the nucleus is not a universal feature of mitosis and does not occur in all cells. Some unicellular eukaryotes (e.g., yeasts) undergo so-called closed mitosis, in which the nuclear envelope remains intact. In closed mitosis, the daughter chromosomes migrate to opposite poles of the nucleus, which then divides in two. The cells of higher eukaryotes, however, usually undergo open mitosis, which is characterized by breakdown of the nuclear envelope. The daughter chromosomes then migrate to opposite poles of the mitotic spindle, and new nuclei reassemble around them.
At a certain point during the cell cycle in open mitosis, the cell divides to form two cells. In order for this process to be possible, each of the new daughter cells must have a full set of genes, a process requiring replication of the chromosomes as well as segregation of the separate sets. This occurs by the replicated chromosomes, the sister chromatids, attaching to microtubules, which in turn are attached to different centrosomes. The sister chromatids can then be pulled to separate locations in the cell. In many cells, the centrosome is located in the cytoplasm, outside the nucleus; the microtubules would be unable to attach to the chromatids in the presence of the nuclear envelope. Therefore, the early stages in the cell cycle, beginning in prophase and until around prometaphase, the nuclear membrane is dismantled. Likewise, during the same period, the nuclear lamina is also disassembled, a process regulated by phosphorylation of the lamins by protein kinases such as the CDC2 protein kinase. Towards the end of the cell cycle, the nuclear membrane is reformed, and around the same time, the nuclear lamina are reassembled by dephosphorylating the lamins.
However, in dinoflagellates, the nuclear envelope remains intact, the centrosomes are located in the cytoplasm, and the microtubules come in contact with chromosomes, whose centromeric regions are incorporated into the nuclear envelope (the so-called closed mitosis with extranuclear spindle). In many other protists (e.g., ciliates, sporozoans) and fungi, the centrosomes are intranuclear, and their nuclear envelope also does not disassemble during cell division.
Apoptosis is a controlled process in which the cell's structural components are destroyed, resulting in death of the cell. Changes associated with apoptosis directly affect the nucleus and its contents, for example, in the condensation of chromatin and the disintegration of the nuclear envelope and lamina. The destruction of the lamin networks is controlled by specialized apoptotic proteases called caspases, which cleave the lamin proteins and, thus, degrade the nucleus' structural integrity. Lamin cleavage is sometimes used as a laboratory indicator of caspase activity in assays for early apoptotic activity. Cells that express mutant caspase-resistant lamins are deficient in nuclear changes related to apoptosis, suggesting that lamins play a role in initiating the events that lead to apoptotic degradation of the nucleus. Inhibition of lamin assembly itself is an inducer of apoptosis.
The nuclear envelope acts as a barrier that prevents both DNA and RNA viruses from entering the nucleus. Some viruses require access to proteins inside the nucleus in order to replicate and/or assemble. DNA viruses, such as herpesvirus replicate and assemble in the cell nucleus, and exit by budding through the inner nuclear membrane. This process is accompanied by disassembly of the lamina on the nuclear face of the inner membrane. |

|
|
|
|
Disease-related dynamics
Disease-related dynamics (W)
Initially, it has been suspected that immunoglobulins in general and autoantibodies in particular do not enter the nucleus. Now there is a body of evidence that under pathological conditions (e.g. lupus erythematosus) IgG can enter the nucleus. |
|
|
|
| |
Nuclei per cell
|
Nuclei per cell
Nuclei per cell (W)
Most eukaryotic cell types usually have a single nucleus, but some have no nuclei, while others have several. This can result from normal development, as in the maturation of mammalian red blood cells, or from faulty cell division. |
|
|
|
Anucleated cells
Anucleated cells (W)
Human red blood cells, like those of other mammals, lack nuclei. This occurs as a normal part of the cells' development. |
|
|
| |
|
An anucleated cell contains no nucleus and is, therefore, incapable of dividing to produce daughter cells. The best-known anucleated cell is the mammalian red blood cell, or erythrocyte, which also lacks other organelles such as mitochondria, and serves primarily as a transport vessel to ferry oxygen from the lungs to the body's tissues. Erythrocytes mature through erythropoiesis in the bone marrow, where they lose their nuclei, organelles, and ribosomes. The nucleus is expelled during the process of differentiation from an erythroblast to a reticulocyte, which is the immediate precursor of the mature erythrocyte. The presence of mutagens may induce the release of some immature "micronucleated" erythrocytes into the bloodstream. Anucleated cells can also arise from flawed cell division in which one daughter lacks a nucleus and the other has two nuclei.
In flowering plants, this condition occurs in sieve tube elements. |

|
|
|
|
|
| |
Evolution
|
Evolution
Evolution (W)
As the major defining characteristic of the eukaryotic cell, the nucleus' evolutionary origin has been the subject of much speculation. Four major hypotheses have been proposed to explain the existence of the nucleus, although none have yet earned widespread support.
The first model known as the "syntrophic model" proposes that a symbiotic relationship between the archaea and bacteria created the nucleus-containing eukaryotic cell. (Organisms of the Archaea and Bacteria domain have no cell nucleus.) It is hypothesized that the symbiosis originated when ancient archaea, similar to modern methanogenic archaea, invaded and lived within bacteria similar to modern myxobacteria, eventually forming the early nucleus. This theory is analogous to the accepted theory for the origin of eukaryotic mitochondria and chloroplasts, which are thought to have developed from a similar endosymbiotic relationship between proto-eukaryotes and aerobic bacteria. The archaeal origin of the nucleus is supported by observations that archaea and eukarya have similar genes for certain proteins, including histones. Observations that myxobacteria are motile, can form multicellular complexes, and possess kinases and G proteins similar to eukarya, support a bacterial origin for the eukaryotic cell.
A second model proposes that proto-eukaryotic cells evolved from bacteria without an endosymbiotic stage. This model is based on the existence of modern planctomycetes bacteria that possess a nuclear structure with primitive pores and other compartmentalized membrane structures. A similar proposal states that a eukaryote-like cell, the chronocyte, evolved first and phagocytosed archaea and bacteria to generate the nucleus and the eukaryotic cell.
The most controversial model, known as viral eukaryogenesis, posits that the membrane-bound nucleus, along with other eukaryotic features, originated from the infection of a prokaryote by a virus. The suggestion is based on similarities between eukaryotes and viruses such as linear DNA strands, mRNA capping, and tight binding to proteins (analogizing histones to viral envelopes). One version of the proposal suggests that the nucleus evolved in concert with phagocytosis to form an early cellular "predator". Another variant proposes that eukaryotes originated from early archaea infected by poxviruses, on the basis of observed similarity between the DNA polymerases in modern poxviruses and eukaryotes. It has been suggested that the unresolved question of the evolution of sex could be related to the viral eukaryogenesis hypothesis.
A more recent proposal, the exomembrane hypothesis, suggests that the nucleus instead originated from a single ancestral cell that evolved a second exterior cell membrane; the interior membrane enclosing the original cell then became the nuclear membrane and evolved increasingly elaborate pore structures for passage of internally synthesized cellular components such as ribosomal subunits. |

|
|
|
|
| |
See also
|
|

|
|
|
|
|
|
The Nucleus (B)
The Nucleus (B)
|
The nucleus is the information centre of the cell and is surrounded by a nuclear membrane in all eukaryotic organisms. It is separated from the cytoplasm by the nuclear envelope, and it houses the double-stranded, spiral-shaped deoxyribonucleic acid ( DNA) molecules, which contain the genetic information necessary for the cell to retain its unique character as it grows and divides.
The presence of a nucleus distinguishes the eukaryotic cells of multicellular organisms from the prokaryotic, one-celled organisms such as bacteria. In contrast to the higher organisms, prokaryotes do not have nuclei, so their DNA is maintained in the same compartment as their other cellular components.
The primary function of the nucleus is the expression of selected subsets of the genetic information encoded in the DNA double helix. Each subset of a DNA chain, called a gene, codes for the construction of a specific protein out of a chain of amino acids. Information in DNA is not decoded directly into proteins, however. First it is transcribed, or copied, into a range of messenger ribonucleic acid (mRNA) molecules, each of which encodes the information for one protein (or more than one protein in bacteria). The mRNA molecules are then transported through the nuclear envelope into the cytoplasm, where they are translated, serving as templates for the synthesis of specific proteins.
The nucleus must not only synthesize the mRNA for many thousands of proteins, but it must also regulate the amounts synthesized and supplied to the cytoplasm. Furthermore, the amounts of each type of mRNA supplied to the cytoplasm must be regulated differently in each type of cell. In addition to mRNA, the nucleus synthesizes and exports other classes of RNA involved in the mechanisms of protein synthesis. |
|
| |
| |
Structural organization of the nucleus
|
Structural organization of the nucleus
Structural organization of the nucleus (B)
No text. |
|
|
|
DNA packaging
DNA packaging (B)
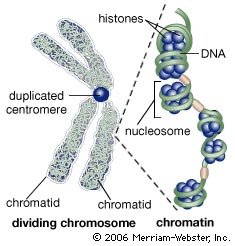
Centromere and chromatids in cell division
During the first stages of cell division, the recognizable double-stranded chromosome is formed by two tightly coiled DNA strands (chromatids) joined at a point called the centromere. During the middle stage of cell division, the centromere duplicates, and the chromatid pair separates. Following cell division, the separated chromatids uncoil; the loosely coiled DNA, wrapped around its associated proteins (histones) to form beaded structures called nucleosomes, is termed chromatin. |
|
|
| |
|
The nucleus of the average human cell is only 6 micrometres (6 × 10−6 metre) in diameter, yet it contains about 1.8 metres of DNA. This is distributed among 46 chromosomes, each consisting of a single DNA molecule about 40 mm (1.5 inches) long. The extraordinary packaging problem this poses can be envisaged by a scale model enlarged a million times. On this scale a DNA molecule would be a thin string 2 mm thick, and the average chromosome would contain 40 km (25 miles) of DNA. With a diameter of only 6 metres, the nucleus would contain 1,800 km (1,118 miles) of DNA.
These contents must be organized in such a way that they can be copied into RNA accurately and selectively. DNA is not simply crammed or wound into the nucleus like a ball of string; rather, it is organized, by molecular interaction with specific nuclear proteins, into a precisely packaged structure. This combination of DNA with proteins creates a dense, compact fibre called chromatin. An extreme example of the ordered folding and compaction that chromatin can undergo is seen during cell division, when the chromatin of each chromosome condenses and is divided between two daughter cells (see below Cell division and growth). |
| |
Nucleosomes: the subunits of chromatin
The compaction of DNA is achieved by winding it around a series of small proteins called histones. Histones are composed of positively charged amino acids that bind tightly to and neutralize the negative charges of DNA. There are five classes of histone. Four of them, called H2A, H2B, H3, and H4, contribute two molecules each to form an octamer, an eight-part core around which two turns of DNA are wrapped. The resulting beadlike structure is called the nucleosome. The DNA enters and leaves a series of nucleosomes, linking them like beads along a string in lengths that vary between species of organism or even between different types of cell within a species. A string of nucleosomes is then coiled into a solenoid configuration by the fifth histone, called H1. One molecule of H1 binds to the site at which DNA enters and leaves each nucleosome, and a chain of H1 molecules coils the string of nucleosomes into the solenoid structure of the chromatin fibre. |
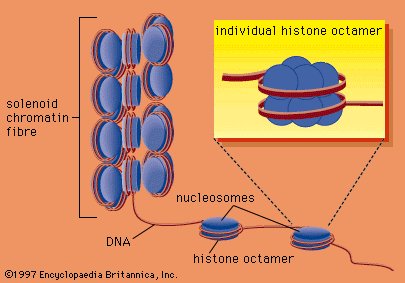
Histone; nucleosome
DNA wrapped around clusters of histone proteins to form nucleosomes, which can coil to form solenoids. |
|
|
| |
|
|
| Nucleosomes not only neutralize the charges of DNA, but they have other consequences. First, they are an efficient means of packaging. DNA becomes compacted by a factor of six when wound into nucleosomes and by a factor of about 40 when the nucleosomes are coiled into a solenoid chromatin fibre. The winding into nucleosomes also allows some inactive DNA to be folded away in inaccessible conformations, a process that contributes to the selectivity of gene expression. |
| |
Organization of chromatin fibre
Several studies indicate that chromatin is organized into a series of large radial loops anchored to specific scaffold proteins. Each loop consists of a chain of nucleosomes and may be related to units of genetic organization. This radial arrangement of chromatin loops compacts DNA about a thousandfold. Further compaction is achieved by a coiling of the entire looped chromatin fibre into a dense structure called a chromatid, two of which form the chromosome. During cell division, this coiling produces a 10,000-fold compaction of DNA. |

|
|
|
|
The nuclear envelope
The nuclear envelope (B)
The nuclear envelope is a double membrane composed of an outer and an inner phospholipid bilayer. The thin space between the two layers connects with the lumen of the rough endoplasmic reticulum (RER), and the outer layer is an extension of the outer face of the RER.
The inner surface of the nuclear envelope has a protein lining called the nuclear lamina, which binds to chromatin and other contents of the nucleus. The entire envelope is perforated by numerous nuclear pores. These transport routes are fully permeable to small molecules up to the size of the smallest proteins, but they form a selective barrier against movement of larger molecules. Each pore is surrounded by an elaborate protein structure called the nuclear pore complex, which selects molecules for entrance into the nucleus. Entering the nucleus through the pores are the nucleotide building blocks of DNA and RNA, as well as adenosine triphosphate, which provides the energy for synthesizing genetic material. Histones and other large proteins must also pass through the pores. These molecules have special amino acid sequences on their surface that signal admittance by the nuclear pore complexes. The complexes also regulate the export from the nucleus of RNA and subunits of ribosomes.
DNA in prokaryotes is also organized in loops and is bound to small proteins resembling histones, but these structures are not enclosed by a nuclear membrane. |

|
|
|
|
| |
Genetic organization of the nucleus
|
Genetic organization of the nucleus
Genetic organization of the nucleus (B)
No text. |
|
|
|
The structure of DNA
The structure of DNA (B)
Several features are common to the genetic structure of most organisms. First is the double-stranded DNA. Each strand of this molecule is a series of nucleotides, and each nucleotide is composed of a sugar-phosphate compound attached to one of four nitrogen-containing bases. The sugar-phosphate compounds link together to form the backbone of the strand. Each of the bases strung along the backbone is chemically attracted to a corresponding base on the parallel strand of the DNA molecule. This base pairing joins the two strands of the molecule much as rungs join the two sides of a ladder, and the chemical bonding of the base pairs twists the doubled strands into a spiral, or helical, shape. |
| |
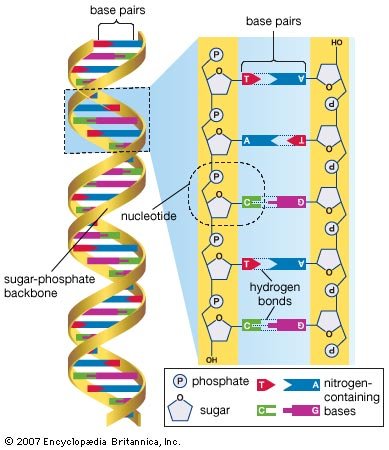
DNA molecule. |
|
|
| |
|
|
| The four nucleotide bases are adenine, cytosine, guanine, and thymine. DNA is composed of millions of these bases strung in an apparently limitless variety of sequences. It is in the sequence of bases that the genetic information is contained, each sequence determining the sequence of amino acids to be connected into proteins. A nucleotide sequence sufficient to encode one protein is called a gene. Genes are interspersed along the DNA molecule with other sequences that do not encode proteins. Some of these so-called untranslated regions regulate the activity of the adjacent genes, for example, by marking the points at which enzymes begin and cease transcribing DNA into RNA (see below Genetic expression through RNA). |

|
|
|
|
Rearrangement and modification of DNA
Rearrangement and modification of DNA (B)
Rearrangements and modifications of the nucleotide sequences in DNA are exceptions to the rules of genetic expression and sometimes cause significant changes in the structure and function of cells. Different cells of the body owe their specialized structures and functions to different genes. This does not mean that the set of genetic information varies among the cells of the body. Indeed, for each cell the entire DNA content of the chromosomes is usually duplicated exactly from generation to generation, and, in general, the genetic content and arrangement is strikingly similar among different cell types of the same organism. As a result, the differentiation of cells can occur without the loss or irreversible inactivation of unnecessary genes, an observation that is reinforced by the presence of specific genes in a range of adult tissues. For example, normal copies of the genes encoding hemoglobin are present in the same numbers in red blood cells, which make hemoglobin, as in a range of other types of cells, which do not. |
| |
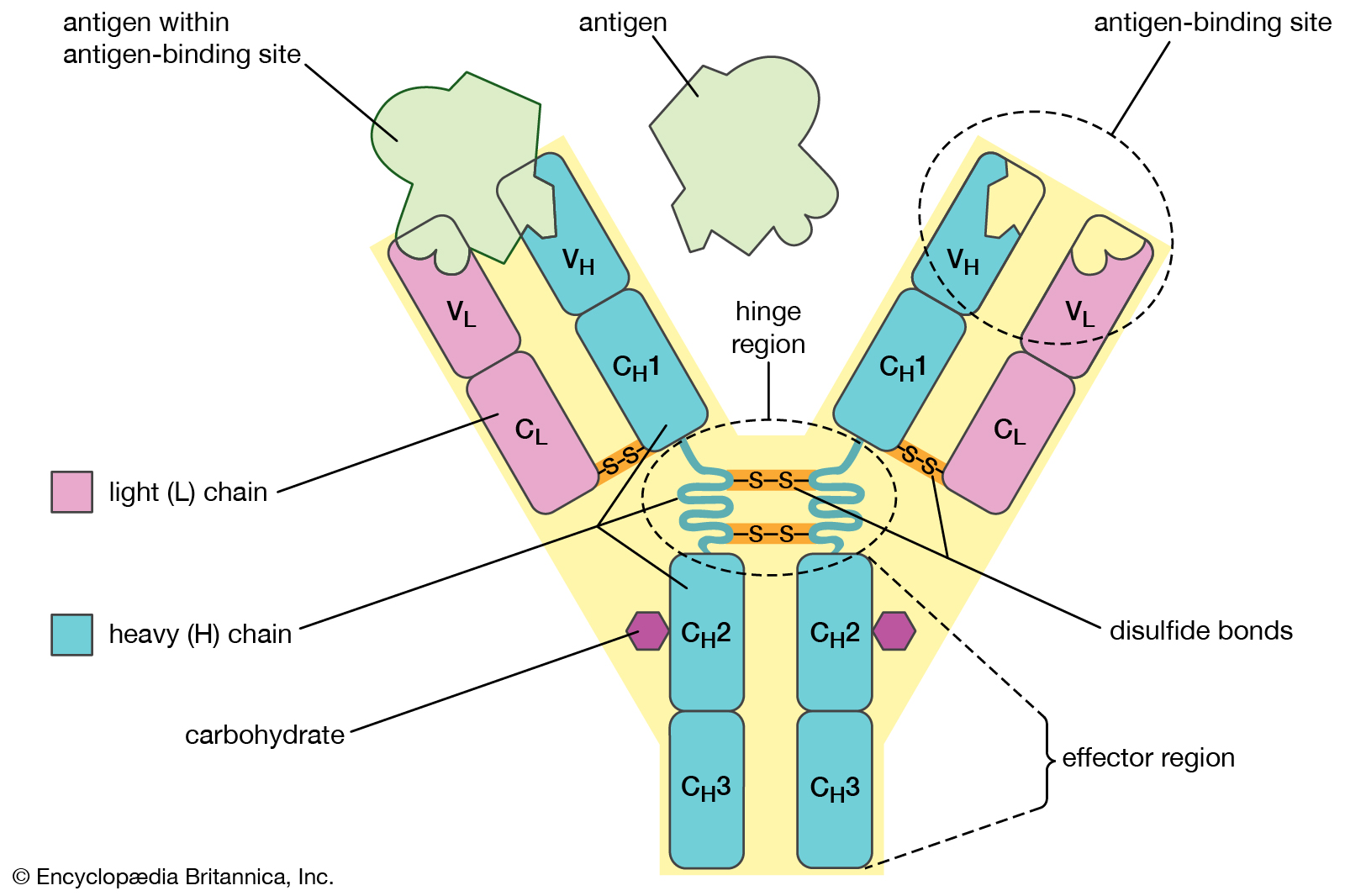
Antibody structure
The structure of an antibody molecule represents the dramatic rearrangements of DNA that occur in the immune systems of mammals. Each antibody contains a light chain and a heavy chain that are encoded by different segments of DNA. These segments are subject to considerable variation and are thus able to produce many different antibodies. |
|
| |
Despite the general uniformity of genetic content in all the cells of an organism, studies have shown a few clear examples in some organisms of programmed, reversible change in the DNA of developing tissues. One of the most dramatic rearrangements of DNA occurs in the immune systems of mammals. The body’s defense against invasion by foreign organisms involves the synthesis of a vast range of antibodies by lymphocytes (a type of white blood cell). Antibodies are proteins that bind to specific invading molecules or organisms and either inactivate them or signal their destruction. The binding sites on each antibody molecule are formed by one light and one heavy amino acid chain, which are encoded by different segments of the DNA in the lymphocyte nucleus. These DNA segments undergo considerable rearrangements, resulting in the synthesis of a great variety of antibodies. Some invasive organisms, such as trypanosome parasites, which cause sleeping sickness, go to great lengths to rearrange their own DNA to evade the versatility of their hosts’ antibody production. The parasites are covered by a thick coat of glycoprotein (a protein with sugars attached). Given time, host organisms can overcome infection by producing antibodies to the parasites’ glycoprotein coat, but this reaction is anticipated and evaded by the selective rearrangement of the trypanosomes’ DNA encoding the glycoprotein, thus constantly changing the surface presented to the hosts’ immune system.
Careful comparisons of gene structure have also revealed epigenetic modifications, heritable changes that occur on the sugar-phosphate side of bases in the DNA and thus do not cause rearrangements in the DNA sequence itself. An example of an epigenetic modification involves the addition of a methyl group to cytosine bases. This appears to cause the inactivation of genes that do not need to be expressed in a particular type of cell. An important feature of the methylation of cytosine lies in its ability to be copied, so that methyl groups in a dividing cell’s DNA will result in methyl groups in the same positions in the DNA of both daughter cells. |

|
|
|
|
| |
Genetic expression through RNA
|
Genetic expression through RNA
Genetic expression through RNA (B)
The transcription of the genetic code from DNA to RNA, and the translation of that code from RNA into protein, exerts the greatest influence on the modulation of genetic information. The process of genetic expression takes place over several stages, and at each stage is the potential for further differentiation of cell types.
As explained above, genetic information is encoded in the sequences of the four nucleotide bases making up a DNA molecule. One of the two DNA strands is transcribed exactly into messenger RNA (mRNA), with the exception that the thymine base of DNA is replaced by uracil. RNA also contains a slightly different sugar component (ribose) from that of DNA (deoxyribose) in its connecting sugar-phosphate chain. Unlike DNA, which is stable throughout the cell’s life and of which individual strands are even passed on to many cell generations, RNA is unstable. It is continuously broken down and replaced, enabling the cell to change its patterns of protein synthesis.
Apart from mRNA, which encodes proteins, other classes of RNA are made by the nucleus. These include ribosomal RNA ( rRNA), which forms part of the ribosomes and is exported to the cytoplasm to help translate the information in mRNA into proteins. Ribosomal RNA is synthesized in a specialized region of the nucleus called the nucleolus, which appears as a dense area within the nucleus and contains the genes that encode rRNA. This is also the site of assembly of ribosome subunits from rRNA and ribosomal proteins. Ribosomal proteins are synthesized in the cytoplasm and transported to the nucleus for subassembly in the nucleolus. The subunits are then returned to the cytoplasm for final assembly. Another class of RNA synthesized in the nucleus is transfer RNA (tRNA), which serves as an adaptor, matching individual amino acids to the nucleotide triplets of mRNA during protein synthesis. |

|
|
|
|
RNA synthesis
RNA synthesis (B)
The synthesis of RNA is performed by enzymes called RNA polymerases. In higher organisms there are three main RNA polymerases, designated I, II, and III (or sometimes A, B, and C). Each is a complex protein consisting of many subunits. RNA polymerase I synthesizes three of the four types of rRNA (called 18S, 28S, and 5.8S RNA); therefore it is active in the nucleolus, where the genes encoding these rRNA molecules reside. RNA polymerase II synthesizes mRNA, though its initial products are not mature RNA but larger precursors, called heterogeneous nuclear RNA, which are completed later (see below Processing of mRNA). The products of RNA polymerase III include tRNA and the fourth RNA component of the ribosome, called 5S RNA.
All three polymerases start RNA synthesis at specific sites on DNA and proceed along the molecule, linking selected nucleotides sequentially until they come to the end of the gene and terminate the growing chain of RNA. Energy for RNA synthesis comes from high-energy phosphate linkages contained in the nucleotide precursors of RNA. Each unit of the final RNA product is essentially a sugar, a base, and one phosphate, but the building material consists of a sugar, a base, and three phosphates. During synthesis two phosphates are cleaved and discarded for each nucleotide that is incorporated into RNA. The energy released from the phosphate bonds is used to link the nucleotides. The crucial feature of RNA synthesis is that the sequence of nucleotides joined into a growing RNA chain is specified by the sequence of nucleotides in the DNA template: each adenine in DNA specifies uracil in RNA, each cytosine specifies guanine, each guanine specifies cytosine, and each thymine in DNA specifies adenine. In this way the information encoded in each gene is transcribed into RNA for translation by the protein-synthesizing machinery of the cytoplasm.
In addition to specifying the sequence of amino acids to be polymerized into proteins, the nucleotide sequence of DNA contains supplementary information. For example, short sequences of nucleotides determine the initiation site for each RNA polymerase, specifying where and when RNA synthesis should occur. In the case of RNA polymerases I and II, the sequences specifying initiation sites lie just ahead of the genes. In contrast, the equivalent information for RNA polymerase III lies within the gene—that is, within the region of DNA to be copied into RNA. The initiation site on a segment of DNA is called a promoter. The promoters of different genes have some nucleotide sequences in common, but they differ in others. The differences in sequence are recognized by specific proteins called transcription factors, which are necessary for the expression of particular types of genes. The specificity of transcription factors contributes to differences in the gene expression of different types of cells. |

|
|
|
|
Processing of mRNA
Processing of mRNA (B)
During and after synthesis, mRNA precursors undergo a complex series of changes before the mature molecules are released from the nucleus. First, a modified nucleotide is added to the start of the RNA molecule by a reaction called capping. This cap later binds to a ribosome in the cytoplasm. The synthesis of mRNA is not terminated simply by the RNA polymerase’s detachment from DNA, but by chemical cleavage of the RNA chain. Many (but not all) types of mRNA have a simple polymer of adenosine residues added to their cleaved ends.
In addition to these modifications of the termini, startling discoveries in 1977 revealed that portions of newly synthesized RNA molecules are cut out and discarded. In many genes, the regions coding for proteins are interrupted by intervening sequences of nucleotides called introns. These introns must be excised from the RNA copy before it can be released from the nucleus as a functional mRNA. The number and size of introns within a gene vary greatly, from no introns at all to more than 50. The sum of the lengths of these intervening sequences is sometimes longer than the sum of the regions coding for proteins.
The removal of introns, called RNA splicing, appears to be mediated by small nuclear ribonucleoprotein particles (snRNP’s). These particles have RNA sequences that are complementary to the junctions between introns and adjacent coding regions. By binding to the junction ends, an snRNP twists the intron into a loop. It then excises the loop and splices the coding regions. |

|
|
|
|
| |
Regulation of genetic expression
|
Regulation of genetic expression
Regulation of genetic expression (B)
Although all the cell nuclei of an organism generally carry the same genes, there are conspicuous differences between the specialized cell types of the body. The source of these differences lies not so much in the occasional modification of DNA, as outlined above, but in the selective expression of DNA through RNA; in particular, it can be traced to processes regulating the amounts and activities of mRNA both during and after its synthesis in the nucleus. |
|
|
|
Regulation of RNA synthesis
Regulation of RNA synthesis (B)
The first level of regulation is mediated by variations in chromatin structure. In order to be transcribed, a gene must be assembled into a structurally distinct form of active chromatin. A second level of regulation is achieved by varying the frequency with which a gene in the active conformation is transcribed into RNA by an RNA polymerase. There is evidence for regulation of RNA synthesis at both these levels—for example, in response to hormone induction. At both levels, protein factors are believed to perform the regulation—for example, by binding to special promoter DNA regions flanking the transcribed gene. |
|
|
|
Regulation of RNA after synthesis
Regulation of RNA after synthesis (B)
After synthesis, RNA molecules undergo selective processing, which results in the export of only a subpopulation of RNA molecules to the cytoplasm. Furthermore, the stability in the cytoplasm of a particular type of mRNA can be regulated. For example, the hormone prolactin increases synthesis of milk proteins in tissue by causing a twofold rise in the rate of mRNA synthesis; but it also causes a 17-fold rise in mRNA lifetime, so that in this case the main cause of increased protein synthesis is the prolonged availability of mRNA. Conversely, there is evidence for selective destabilization of some mRNA—such as histone mRNA, which is rapidly broken down when DNA replication is interrupted. Finally, there are many examples of selective regulation of the translation of mRNA into protein. |
|
|
|

|
|
|
|
|
|

